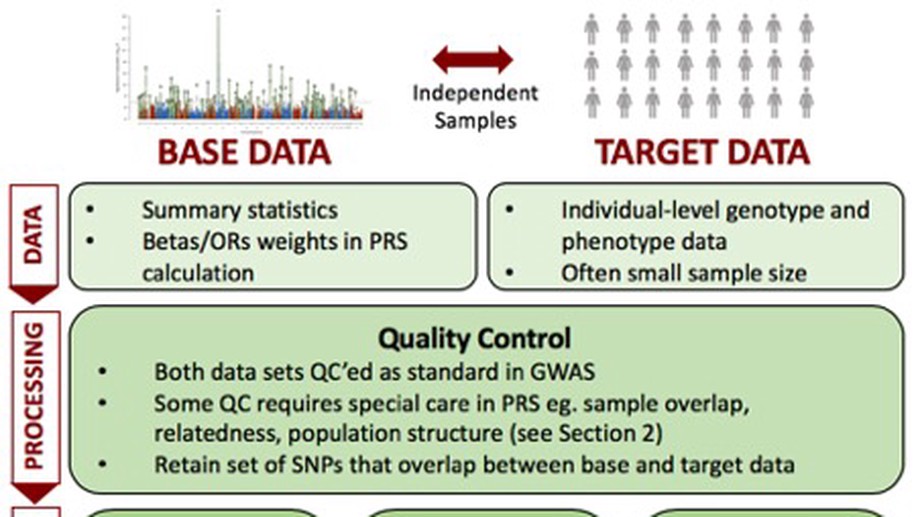
Background: Polygenic risk score (PRS) analyses have become an integral part of biomedical research, exploited to gain insights into shared aetiology among traits, to control for genomic profile in experimental studies, and to strengthen causal inference, among a range of applications. Substantial efforts are now devoted to biobank projects to collect large genetic and phenotypic data, providing unprecedented opportunity for genetic discovery and applications. To process the large-scale data provided by such biobank resources, highly efficient and scalable methods and software are required.
Results: Here we introduce PRSice-2, an efficient and scalable software for automating and simplifying polygenic risk score analyses on large-scale data. PRSice-2 handles both genotyped and imputed data, provides empirical association P-values free from inflation due to overfitting, supports different inheritance models and can evaluate multiple continuous and binary target traits simultaneously. We demonstrate that PRSice-2 is dramatically faster and more memory-efficient than PRSice-1 and alternative polygenic score software, LDpred and lassosum, while having comparable predictive power.
Conclusion: PRSice-2’s combination of efficiency and power will be increasingly important as data sizes grow and as the applications of PRS become more sophisticated; for example, when incorporated into high-dimensional or gene-set based analyses. PRSice-2 is written in C++, with an R script for plotting, and is freely available for download from
http://PRSice.info